Synthesizing new proteins – the constructing blocks of organic life – is a scientific discipline of immense potential, and a newly developed AI mannequin guarantees to create directions for brand spanking new proteins means past these present in nature.
Scientists within the US have used EvolutionaryScale Mannequin 3 (ESM3) to synthesize a brand new protein referred to as esmGFP (inexperienced fluorescent protein), which solely shares 58 % of its materials with its closest pure relative tagRFP.
That is the equal of 500 million years of evolution being processed by AI, the analysis group estimates, and it opens the best way to creating custom-made proteins that may be designed for particular makes use of, or unlocking extra features from present proteins.
“More than three billion years of evolution have produced an image of biology encoded into the space of natural proteins,” write the researchers, led by Thomas Hayes, founding father of EvolutionaryScale in New York, of their printed paper.
“Here we show that language models trained at scale on evolutionary data can generate functional proteins that are far away from known proteins.”
I’m so excited to share what we’ve been engaged on @EvoscaleAI. ESM3 is a multimodal generative masked language mannequin for programming biology. Right here’s a brief thread on the structure behind ESM3. 🧵https://t.co/jldHYRAPNy
— Thomas Hayes (@THayes427) June 25, 2024
ESM3 was skilled on a formidable 3.15 billion protein sequences (the order of amino acids in a protein), 236 million protein constructions (their 3D shapes), and 539 million protein annotations (descriptive labels).
By recognizing patterns in these huge troves of information, the AI mannequin can perceive what works and what does not in protein constructing and performance – in the identical means that ChatGPT can compose a brand new poem that rhymes after studying tens of millions of poems written by people.
What makes esmGFP additional particular is that it really works: it is fluorescent similar to its relative tagRFP. Fluorescent proteins give some ocean organisms their glow, and their use as markers have large significance in drugs and biotechnology.
“We chose the functionality of fluorescence because it is difficult to achieve, easy to measure, and one of the most beautiful mechanisms in nature,” the group writes.
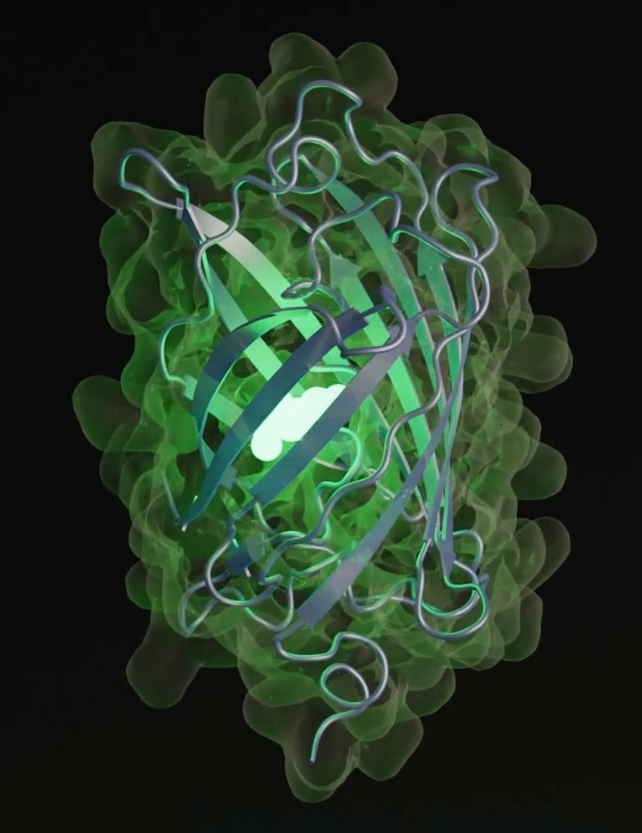
The AI takes away a variety of the trial and error in protein synthesis, whereas including the power to discover distant from proteins we at present find out about.
“Proteins can be seen as existing within an organized space where each protein is neighbored by every other that is one mutational event away,” write the researchers. “The structure of evolution appears as a network within this space, connecting all proteins by the paths that evolution can take between them.”
For evolution to happen, the group says every protein should develop into the subsequent one with out the system of which it’s a half dropping its total performance. A language mannequin acknowledges proteins on this house.
Proteins designed by ESM3 nonetheless must be validated, synthesized, and examined, which takes time, however the group is assured of creating additional progress right here. Within the not-too-distant future we could possibly be producing proteins for every little thing from medicines to biomaterials simply with some intelligent AI prompting.
“Protein language models do not explicitly work within the physical constraints of evolution, but instead can implicitly construct a model of the multitude of potential paths evolution could have followed,” the researchers clarify.
The analysis has been printed in Science.

